

To publish your data on portals like GBIF
data can be provided in the ABCD
standard using
BioCASe
 for mapping your data (see below). Use the
ABCD package to convert the data into a format prepared for BioCASe. Configure
the database and map the fields according to the examples.
for mapping your data (see below). Use the
ABCD package to convert the data into a format prepared for BioCASe. Configure
the database and map the fields according to the examples.
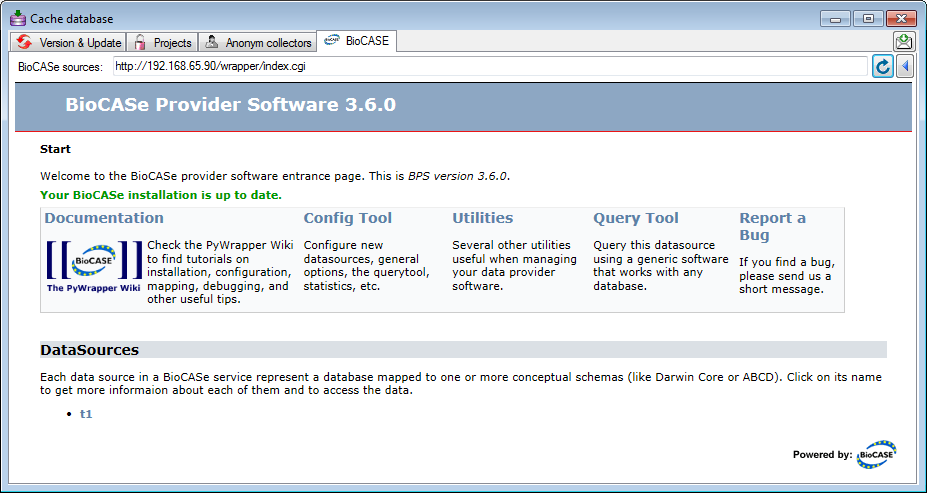
For details about the BioCASe software please see the provided documentation. DiversityCollection transfers the data for every project in a separate schema. As BioCASe so far is unable to handle schemata in Postgres additional views for the package ABCD are created in the schema public and you need to provide a single database for every project.
To provide the data for BioCASe you need to
transfer the data from DiversityCollection together with all auxiliary sources
(Taxa, References, Gazetteer) depending on your data from the
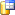 SQL-Server database to the
SQL-Server database to the
 SQL-Server cache database and from there to
the
SQL-Server cache database and from there to
the
 Postgres cache database. In the Postgres
database you need the package
Postgres cache database. In the Postgres
database you need the package
 ABCD for mapping your data to BioCASe.
ABCD for mapping your data to BioCASe.
| Database | Table | Column | ABCD | Node |
| DiversityAgents | Agent | |||
| DiversityAgents | AgentContactInformation | |||
| DiversityAgents | AgentContactInformation | Address | ||
| DiversityProjects | Project | |||
 Settings as shown below as children of the
setting ABCD. The following ABCD topics are taken from the settings in
DiversityProjects:
Settings as shown below as children of the
setting ABCD. The following ABCD topics are taken from the settings in
DiversityProjects:
For further information about the configuration of the settings in DiversityProjects, please see the manual for this module.
The DatasetGUID in ABCD is taken from the stable
identifier defined in the module DiversityProjects for the database in
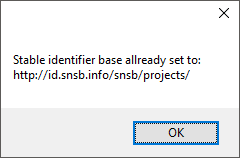
combination with the ID of the project. To set the stable identifier choose
Administration -
 Stable identifier from the menu in
DiversityProjects and enter the basic path (e.g. see below). This stable
identifier can only be set once. After it is set you will get a message as
shown below.
Stable identifier from the menu in
DiversityProjects and enter the basic path (e.g. see below). This stable
identifier can only be set once. After it is set you will get a message as
shown below.
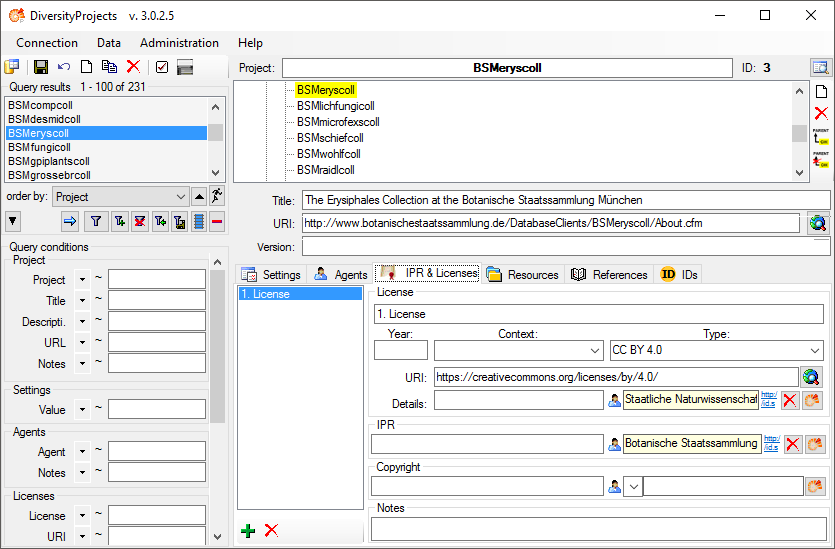
The license information for ABCD are taken from
the first entry in the
 IPR & Licenses section in DiversityProjects
(see below) where the first entry is the one entered first.
IPR & Licenses section in DiversityProjects
(see below) where the first entry is the one entered first.
The citation for the project follows the form <Authors> (<Publication_year>). <Title>. [Dataset]. Data Publisher: <Data_center_name>. <URI>.
The <Authors> are
taken from the
 Agents section
(see below) where all agents with the role Author
are included according to their sequence in the list and separated by ";".
Agents section
(see below) where all agents with the role Author
are included according to their sequence in the list and separated by ";".
The <Publication_year>
is the year of the publication i.e. the current year. The
<Title> corresponds to the Title of the
project. The <Data_center_name> is taken from the
 Agents
section. Here the first agent with the role Publisher
is used. The <URI> is taken from the field URI of
the citation with the type BioCASe (GFBio) in the
Agents
section. Here the first agent with the role Publisher
is used. The <URI> is taken from the field URI of
the citation with the type BioCASe (GFBio) in the
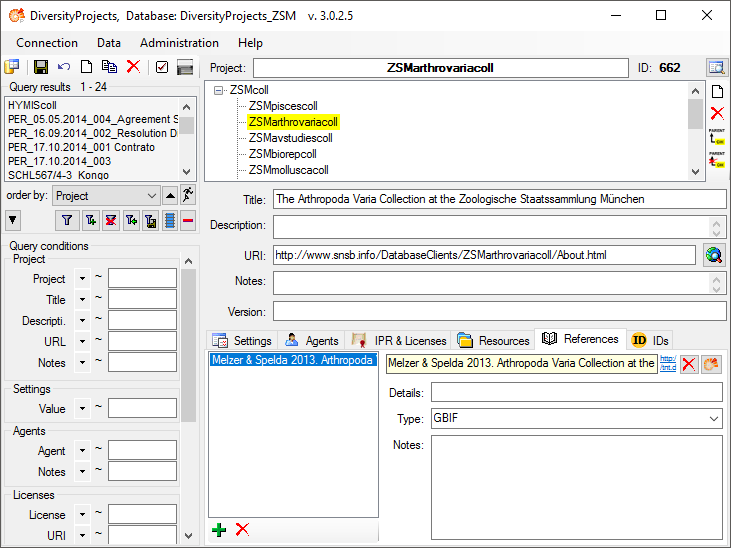
 Citations &
References section (see below).
Citations &
References section (see below).

The IsoType for a country is taken from corresponding information in the module DiversityGazetteer. To ensure these information is available in the cache database, insert a source for a gazetteer project containing this information (see below).
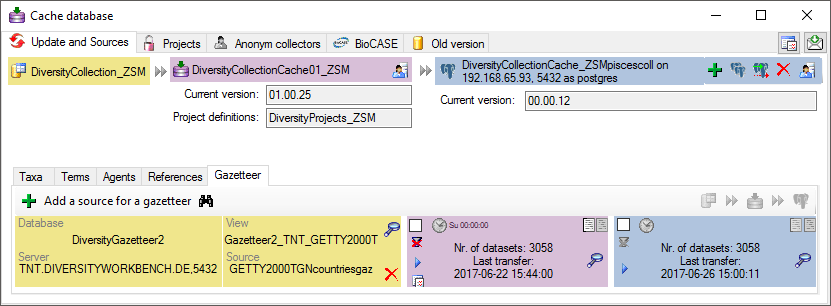
The ABCD schema provided with this software is NOT including information from the module DiversityTaxonNames, so sources for Taxa are not needed by default. If for your own needs you decide to add additional data for the identifications from the module DivesityTaxonNames, ensure that the data corresponding to the names in your identifications are transferred from DiversityTaxonNames (see below).
The package ABCD providing the objects for mapping your data for BioCASE contains objects in the schema public. Therefore you need one database for every project you want to provide for BioCASE and the sources mentioned above have to be transferred in each of these databases.
To
provide the data for BioCASE you need to transfer all auxiliary sources as
described above (Taxa, References, Gazetteer) depending on your data and the
project data themselves from the SQL-Server databases
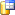 to the SQL-Server cache database
to the SQL-Server cache database
 and from there to the Postgres cache
database
and from there to the Postgres cache
database
 . The package itself needs a further
transfer step
. The package itself needs a further
transfer step
 if data have been
changed after the creation of the package as the parts of the data in the Postgres database
are imported into tables or materialized views according to the specifications of ABCD.
if data have been
changed after the creation of the package as the parts of the data in the Postgres database
are imported into tables or materialized views according to the specifications of ABCD.
